Yeast Adaptation Study Finds Diploids Evolve More Slowly than Haploids
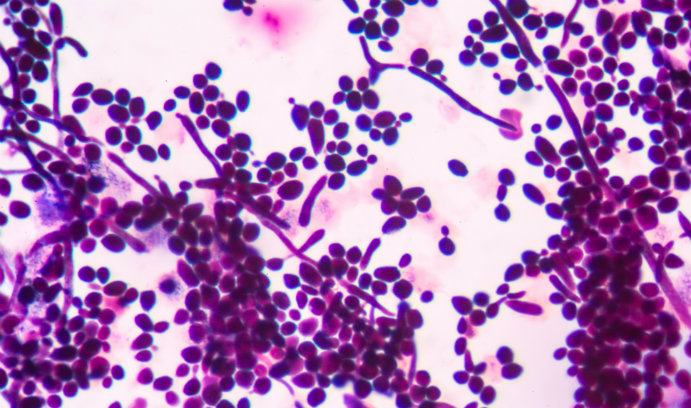
An image of budding yeast cells (Image credit: iStock/toeytoey2530)
Experimental evolution is a good way to enhance our current understanding of how genomes—or sets of chromosomes in an organism’s cells—evolve and the role of individual mutations in adaptation.
Organisms differ in ploidy, or how many copies of the genome they carry in their cells. For example, says Gregory Lang, assistant professor of biological sciences, humans have two copies of our genome in each cell—one from the mom and one from the dad. Bacteria have one copy of their genome in each cell. The common strawberry has eight copies. In other words, humans are diploid, bacteria are haploid, and strawberries are octoploid.
Understanding the influence of ploidy on evolution is only possible through experimental evolution organisms such as yeast. Not only can yeast undergo as many as ten generations in a 24-hour period, it can also be stably maintained at different ploidies.
Lang, along with graduate student Daniel A. Marad and post-doc Sean W. Buskirk, set out to answer a basic question: How do the rates of adaptation differ between haploid and diploid organisms? They found that diploids, with two copies of the genome, evolve more slowly than haploids, which have only one copy. They also found that the beneficial mutations diploids pick up look different compared to what is seen in haploids.
Their results have been published in a paper in Nature Ecology & Evolution called “Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids.”
To understand these dynamics, the team measured the rate of adaptation for 48 diploid populations through 4,000 generations of the yeast Saccharomyces cerevisiae and compared these results to previously evolved haploid populations. They sequenced two clones each from 24 populations after 2,000 generations and performed whole-genome whole-population time-course sequencing on two populations.
“Using a powerful combination of experimental evolution and whole-genome sequencing, we determined the rate of adaptation and the types of mutations that arise in populations of yeast that are identical except for the number of copies of their genome,” says Lang.
“We show that diploids adapt more slowly than haploids, that ploidy alters the spectrum of beneficial mutations, and that the prevalence of homozygous mutations depends on their genomic position,” says Marad. “In addition, we validate haploid-specific, diploid-specific and shared mutational targets by reconstruction.”
According to Lang, evolutionary biology has a rich history, with many classical theories still in need of experimental tests. One theory, known as Haldane's sieve postulates that beneficial mutations that are recessive, or have no selective benefit when present in only one copy in a diploid, are unable to increase in frequency in diploid populations.
Lang and his colleagues tested Haldane's sieve by taking beneficial mutations that arose in haploids and moving them individually into a different context: diploids, to see if they have a selective advantage. Consistent with Haldane's sieve they found that beneficial mutations that arise in diploids are not recessive, but that most beneficial mutations in haploids are.
“Collectively, this work fills a gap in our understanding of how ploidy impacts adaptation, and provides empirical support for the hypothesis that diploid populations have altered access to beneficial mutations,” says Marad.
Lang says a key takeaway from the paper is that ploidy, which varies considerably in the natural world, has a significant effect on how genomes can and will evolve.
This work was supported by the Charles E. Kaufman Foundation of The Pittsburgh Foundation.
Posted on: