Wonpil Im Wins Prestigious German Research Honor
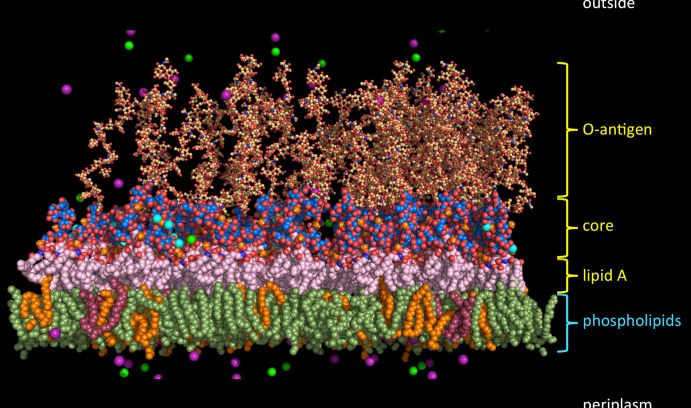
An example of the molecular complexity of the outer cell membrane of a Gram-negative bacterium is shown in this illustration of a typical E. coli outer membrane. (Image courtesy of Wonpil Im)
Bacteria have a formidable enemy in Wonpil Im.
Im, the Presidential Endowed Chair in Health and Professor of Biological Sciences and Bioengineering, uses computational biophysics to learn how antibiotics permeate bacterial membranes and target bacteria for destruction.
His research group has developed CHARMM-GUI, an open-access research program that simulates complex biomolecular systems more simply and more precisely than previously possible. The tool is becoming increasingly valuable as more bacteria develop resistance to antibiotics.
Im’s research recently got a boost when he received the prestigious Friedrich Wilhelm Bessel Research Award from the Alexander von Humboldt Foundation, an organization of the German government that promotes collaboration between scientists from Germany and abroad.
The Bessel Award is named for Friedrich Wilhelm Bessel, a 19th-century German astronomer and mathematician. It will support research that Im is conducting with Ulrich Kleinekathöfer, professor of theoretical physics, and Mathias Winterhalter, an experimental biophysicist, both at Jacobs University in Bremen, Germany.
“With the Humboldt Foundation awarding only 20 Bessel Research Awards each year, this award is evidence of Wonpil’s accomplishments in the field and his international colleagues’ eagerness to work with him,” said Alan J. Snyder, vice president and associate provost for research and graduate studies at Lehigh.
In their project, Im, Kleinekathöfer and Winterhalter will use computational biophysics to model bacterial outer membrane proteins and the transport of molecules, especially antibiotic molecules. Im will use a specialized CHARMM-GUI module to build a simulation of the complex outer membrane of Gram-negative bacteria.
Gram-negative bacteria are more resistant to antibiotics than Gram-positive bacteria because their impenetrable cell wall is made up of both an inner and outer membrane. Diseases caused by Gram-negative bacteria include cholera, typhoid and meningitis, as well as infections caused by the bacterium E. coli.
“The work we are doing is aligned with one of the top drug development priorities identified by the European Commission’s ‘New Drugs for Bad Bugs’ initiative,” said Im. “These priorities include translocation, which focuses on understanding the molecular basis of bacterial cell wall permeability.
“Of particular concern is how antibiotic substances are transported across bacterial cell walls into the pathogens with the help of highly specialized transport proteins. We don’t really know the mechanics of how molecules penetrate the outer bacterial membrane. By understanding this process thoroughly, researchers could more easily predict what kinds of molecular structures could target specific bacterial proteins and kill the cell.”
Im’s group recently learned how to use lipopolysaccharide, a simple phospholipid, to mimic the outer membrane of E. coli. It was the first major step for his lab to simulate a Gram-negative pathogen for drug discovery. Im’s CHARMM-GUI can model lipopolysaccharide structures’ various bacteria in less than 10 minutes.
Im’s ultimate goal is to use CHARMM-GUI to model complex biomolecular systems that will further scientific understanding of the structure and functions of 10 different superbugs.
“I hope that widespread access to this free graphical user interface will enable researchers worldwide to model any number of bacterial cells efficiently,” he said. “This will push the boundaries of our modeling capabilities and lead to a greater understanding of how these complex systems actually work.”
Im’s work is also supported by two programs of the National Science Foundation: the Molecular and Cellular Biosciences program and the Advances in Bio Informatics program.
Story by Lori Friedman
Posted on:





