CHARMM-GUI Provides a Close-up Look at Bacteria
Antibiotics fight bacterial infections by either killing bacteria or impeding their ability to grow and multiply.
Sometimes, though, they don’t work.
Over time, certain types of bacteria—including Gram-negative bacteria like E. coli and Pseudomonas aeruginosa—change their behavior to overcome the impact of the drugs designed to destroy them. Infections caused by these drug-resistant bacteria are difficult to treat and can be deadly.
“Eventually we will have better ideas about antibiotics and how to design antibiotics,” says Wonpil Im, professor of biological sciences and bioengineering and Presidential Endowed Chair in Health Science and Engineering.

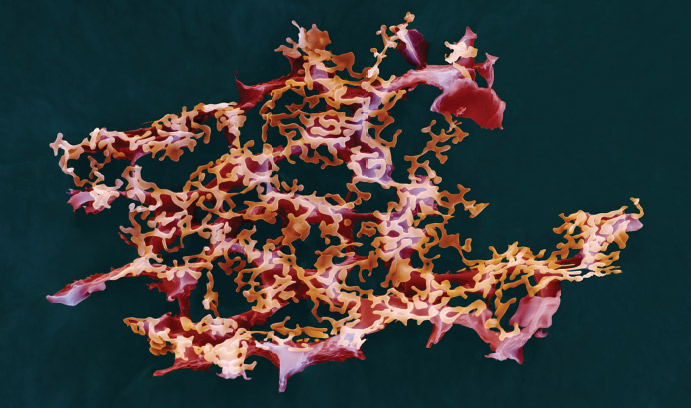
Im is attempting to advance that process by providing researchers with a better understanding of bacteria through CHARMM-GUI (http://www.charmm-gui.org), an open-access research program he and his team have developed. CHARMM-GUI models complex biomolecular systems in just minutes or hours, allowing researchers to produce models of, for example, the properties of the simple phospholipid lipopolysaccharide, an important component of the outer membrane of Gram-negative bacteria. These models allow for a closer look at the molecules that experimental research alone cannot provide.
When scientists discover something new about a biological process through experimental research, says Im, they continue to ask questions about that process: What are the key players? What proteins and molecules are involved? What are the structures and functions of these molecules? How do they move and interact?
“That is sometimes extremely challenging to study solely by experiment,” says Im. “Doing modeling and simulation complements experiments and also makes our understanding better for certain biological questions. [Researchers] are trying to understand the function and dynamics of these biomolecules at the molecular level, at the atomic level, by doing this kind of modeling and simulation.”
Im runs quite possibly the only lab in the world that can systematically build models of complex systems like bacterial outer membranes.
Reducing Barriers
Martin Karplus, recipient of the 2013 Nobel Prize in Chemistry, has led the development of CHARMM (Chemistry at HARvard Macromolecular Mechanics), a molecular simulation program that primarily targets biological systems. CHARMM is widely used, but, says Im, “there is a steep learning curve.”
To help make CHARMM more accessible to users, Im and his team in 2006 started to develop CHARMM-GUI, a graphical user interface for CHARMM. Today, CHARMM-GUI can be used for all major simulation programs, including CHARMM, NAMD, GROMACS, AMBER, GENESIS, LAMMPS, Desmond, OpenMM, and CHARMM/OpenMM simulation programs mostly based on the CHARMM force fields.
Im has granted open access to CHARMM-GUI so that researchers around the world can produce better, higher-level research.
“The main purpose is to reduce the barrier,” says Im. “[Building] very complex simulation programs [without CHARMM-GUI] could take several days or several weeks, sometimes several months.”
Indeed, the initial modeling of a single lipopolysaccharide molecule for CHARMM-GUI took Im’s team six months. At this point, they have modeled most known lipopolysaccharides from various Gram-negative bacteria. Thanks to that extensive, time-consuming work, CHARMM-GUI users can build a molecule in just minutes, or an outer membrane in a matter of hours. This can help speed along the process of finding answers in the fight against bacteria, Im says.
“People can actually spend more time with scientific questions, [so] overall as a community we could do better research,” he says.
Im likens CHARMM-GUI to a “computational microscope.”
“Now, instead of looking at organelles or cells that are moving, we are looking at individual atoms [and] how the molecules are moving and interacting as a function of time,” he says. “These molecules are not visible at [the microscopic] level. There is no technique you can [use to] actually see their motions clearly. This modeling and simulation actually visualizes things. It provides a way of changing the way of your thinking. By just looking at the system, you may learn things that you didn’t even think about before. That kind of thing helps experimental [researchers].”
The Mystery of Translocation
In order to do its job, an antibiotic must first penetrate the outer membrane of a bacterial cell through a process called translocation. Scientists know that antibiotics are able to do this—after all, they work. The mystery here is how they do it.
Understanding translocation, says Im, is essential to the development of effective drugs.
“The target of an antibiotic is not on the surface of the bacteria—it's on the inside,” he says. “So the drug has to go through the outer membrane [and] sometimes an inner membrane to target specific molecules to kill the bacteria. And we really have no idea how that’s happening.”
Ultimately, Im hopes CHARMM-GUI can help researchers understand the structure and functions of a variety of pathogenic bacteria, which can then potentially help in the design of more effective antibiotics.
Mutually Beneficial Partnerships
Germany’s Alexander von Humboldt Foundation recently recognized Im with the prestigious Friedrich Wilhelm Bessel Research Award, which promotes collaboration between scientists in Germany and abroad. The award supports Im’s research with two researchers at Jacobs University in Bremen, Germany: Ulrich Kleinekathöfer, professor of theoretical physics, and experimental biophysicist Mathias Winterhalter. The team uses computational biophysics to model bacterial outer membrane proteins and the transport of molecules.
At Lehigh, Im collaborates with Marcos Pires, assistant professor of chemistry, whose work focuses on the chemical biology of bacterial cell surfaces. To alert the immune system to the presence of bacteria and engage it in the fight against infection, Pires and his team manufacture molecules that stick to the surface of bacteria and attract the immune system. They then monitor the molecules to see what happens when they are introduced to antibodies. But, says Pires, they can’t get a complete picture of what’s happening on the bacterial surface with just an experimental approach.
“There are very few studies out there, simply because it’s really hard to make these measurements, and it’s really hard to put together the surface of the bacteria because it’s so very complex,” says Pires. “[Im’s] got a system, so he’s trying to leverage that to ask these questions. If you have an antibiotic that’s known to come through [the bacterial membrane], he can sort of simulate and see which way it’s coming through. If you have something that’s known not to come through, he can simulate it and try to tease out what controls whether something gets through or not.”
The speed with which Im is able to generate bacterial surfaces helps speed up the process of Pires’ research, and Pires’ experimental data helps Im determine the accuracy of his simulations. If Pires’ team notices a trend in their experimental data, Im can create a simulation to try to explain it.
The two share and benchmark their data, with the goal of developing a predictive algorithm. If they’re successful, says Pires, they will be able to predict whether a particular molecule will or will not be able to pass through a bacterial membrane.
“If you have a drug-like molecule, [we can tell you] if you put this here and here, that’s likely to make it go in. That’s a huge goal,” says Pires.
A predictive algorithm like this could give researchers what they seek in the fight against drug-resistant bacteria: a win.
“It’s an exciting problem,” says Im. “It’s challenging. … [But if] we can actually understand, we might be actually able to better design antibiotics.”
Posted on:




